ANTH domain
| ANTH domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
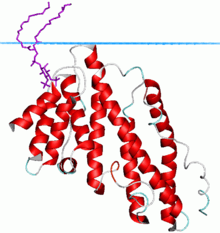 Clathrin assembly lymphoid myeloid leukemia (CALM) protein | |||||||||
| Identifiers | |||||||||
| Symbol | ANTH | ||||||||
| Pfam | PF07651 | ||||||||
| InterPro | IPR011417 | ||||||||
| OPM superfamily | 39 | ||||||||
| OPM protein | 1hfa | ||||||||
| CDD | cd03564 | ||||||||
| |||||||||
The ANTH domain is a membrane binding domain that shows weak specificity for PtdIns(4,5)P2. It was found in AP180 (a.k.a. CALM) endocytotic accessory protein that has been implicated in the formation of clathrin-coated pits. The domain is involved in phosphatidylinositol 4,5-bisphosphate binding and is a universal adaptor for nucleation of clathrin coats.[1][2]
Its structure is a solenoid of 9 helices. The PtdIns(4,5)P2 binding residues are spread over several helices at the tip of the structure. The PtdIns(4,5)P2 binding sequence is Kx9Kx(K/R)(H/Y).
An ANTH domain is also found in HIP1 and HIP1R, and the PtdIns(4,5)P2 binding sequence is conserved. More information is found on endocytosis.org.
Human proteins containing this domain
References
- ↑ de Camilli P, McMahon HT, Peter BJ, Stahelin RV, Cho W, Long F, Murray D (2003). "Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains". J. Biol. Chem. 278 (31): 28993–9. doi:10.1074/jbc.M302865200. PMID 12740367.
- ↑ Payne GS, Duncan MC (2003). "ENTH/ANTH domains expand to the Golgi". Trends Cell Biol. 13 (5): 211–5. doi:10.1016/S0962-8924(03)00076-X. PMID 12742163.
Further reading
- Ford MG, Pearse BM, Higgins MK, et al. (February 2001). "Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes". Science. 291 (5506): 1051–5. doi:10.1126/science.291.5506.1051. PMID 11161218.
External links
- UMich Orientation of Proteins in Membranes protein/pdbid-1hfa - Calculated spatial position of ANTH domain of CALM protein in membrane
This article is issued from Wikipedia - version of the 5/14/2016. The text is available under the Creative Commons Attribution/Share Alike but additional terms may apply for the media files.