C12orf42
Chromosome 12 Open Reading Frame 42 (C12orf42) is a protein-encoding gene in Homo sapiens.
Gene
Locus
The genomic location for this gene is as follows: starts at 103,237,591 bp and ends 103,496,010 bp.[1] The cytogenetic location for C12orf42 is 12q23.2. It is located on the negative strand[2]
mRNA
Fifteen different mRNAs are made by transcription: fourteen alternative splice variants and one unspliced form.[2]
Protein
The protein released by this gene is known as uncharacterized protein C12orf42.[1] There are three isoforms for this protein produced by alternative splicing. The first isoform is a conical sequence. The second Isoform differs form the first in that it doesn't contain 1-95 aa in its sequence. The third isoform differs from the conical sequence in two ways:[5]
- 87-107 aa is the sequence GSHHGQATQKLQGAMVLHLEE instead of VFPERTQNSMACKRLLHTCQY
- the entire sequence 108-360 aa doesn't exist in this isoform
Secondary structure
C12orf42 protein takes on several secondary structures, such as: alpha helices, beta sheets, and random coils. C12orf42 protein is a soluble.[6] Proteins that are soluble have a hydrophilic outside and hydrophobic interior .[7] Proteins with this type of structure are able to freely float inside a cell, due to the liquid composition of the cytosol.
Subcellular location
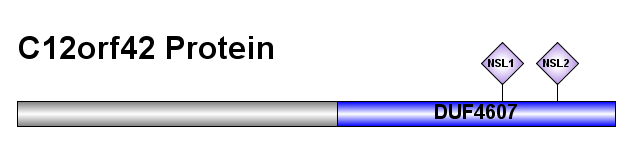
C12orf42 is an intracellular protein. This is known by the lack of transmembrane domains or signal peptides. This suggests that it is predicted to be a nuclear protein, given the nuclear localization signal (NSL) found: PRDRRPQ at 292 aa and a bipartite KRLIKVCSSAPPRPTRR at 325 aa.
Post-translation modification
Predicted post-translation modification sites are seen below in the table. Nuclear proteins are known for having phosphorylation, acetylation, sumoylation, and O-GlcNAc as types of modifications:[8]
- Phosphorylation affects proteins-protein interaction and the stability of the protein.[8]
- Acetylation promotes protein folding and improves stability.
- Sumoylation is involved in nuclear-cytosolic transport and DNA repair.[8]
- Glycosylation (known as O-GlcNAc while in the nucleus) promotes protein folding and stability.
| Type of Modification | Amino Acid Position |
| Phosphorylation | Ser44,Ser47,Ser58,Ser74,Ser113,Ser115,Ser118,Ser123,Ser130,Ser134,Ser135,Ser205,Ser210,Ser217,
Ser226,Ser238, Ser302,Thr17,Thr45,Thr145,Thr150,Thr228, Thr240,Thr240,Thr291,Thr339,Thr344,Tyr124[9] |
| Acetylation | Ser2[10] |
| Sumoylation | IPIVS32-36[11] |
| O-GlcNAc | Thr45,Ser58,Ser130,Ser135,Ser205,Ser210,
Ser217,Thr339[12] |
Expression
Tissue profiles
Microarray data shows expression of the C12orf42 gene in different tissues throughout the human body. There is high expression in the lymph node, spleen, and thymus.[13] There is significant expression in the brain, bladder, epididymis, and the helper T cell.[13] Therefore, there is statistically significant expression of C12orf42 gene throughout the nervous system, immune system, and male reproductive system.
In situ hybridization
The table below shows the areas in the mouse brain where C12orf42 is expressed. The gene name for the mouse is 1700113H08Rik, it is the human homolog of C12orf42.[14] Area one and two of the brain manages body and skeletal movement. Areas three and four in the brain are for sensory functions; area four specializes in perception of smell. Area five in the brain functions in emotional learning and memory.
Homology
Paralog
C12orf42 gene has only one other member in its gene family, this gene is known as Neuroligin 4, Y linked gene (NLGN4Y).[16]
Orthologs
C12orf42 orthologs are mostly mammals. One exception that was found is the Pelodiscus Sinensis or more commonly known as the Chinese soft-shell turtle.
Conserved domain structure
The domain structure that is most important is DUF4607, it is conserved in the Eutheria clade in the Mammalia class. The order that it is conserved in is as follows: Artiodactyla, Carnivora, Chiroptera, Lagomorpha, Perissodactyla, Primates, Proboscidea, and Rodentia.
| Mammalia Class | genus | species | common name | Date of divergence | accession # | seq length | seq ident | seq similar | Taxa/Paralog |
| 1 | Homo | H.Sapiens | Human | --------------- | NP_001157710 | 256aa | 89.80% | ***** | Paralog |
| 2 | Papio | P.Anubis | Olive Baboon | 27.3 mya | XP_009179812 | 353aa | 90% | 93% | Order Primate |
| 3 | Ovis | O. Aries | Sheep | 95.0 mya | XP_012026728 | 177aa | 64.00% | 68% | Order Artiodactyla |
| 4 | Oryctolagus | O. Cuniculus | European Rabbit | 90.1 mya | XP_008255213 | 346aa | 61% | 70% | Order Lagomorpha |
| 5 | Equus | E. Caballus | Horse | 95.0 mya | XP_005606608 | 293aa | 60% | 68% | Order Perissodactyla |
| 6 | Orcinus | O. Orca | Killer Whale | 95.0 mya | XP_012388341.1 | 322aa | 59% | 67% | Order Cetacea |
| 7 | Galeopterus | G.Variegatus | Sunda flying lemur | 83.0 mya | XP_008574769.1 | 289aa | 56% | 64% | Order Dermoptera |
| 8 | Trichechus | T.Manatus | West Indian Manatee | 102.0 mya | XP_012410246 | 348aa | 55% | 63% | Order Sirenia |
| 9 | Loxodonta | L. Africana | African bush elephant | 102.0 mya | XP_010599824 | 288aa | 54% | 62% | Order Proboscidea |
| 10 | Pteropus | P.Alecto | Black flying fox | 95.0 mya | ELK10322.1 | 300aa | 52% | 64% | Order Chiroptera |
| 11 | Condylura | c.Cristata | Star-nose mole | 95.0 mya | XP_004676538.1 | 306aa | 52% | 64% | Order Insectivora |
| 12 | Ailuropoda | A. Melanoleuca | Giant Panda | 95.0 mya | XP_011218367.1 | 305aa | 51% | 61% | Order Carnivora |
| 13 | orycteropus | O. Afer | Aardvarks | 102.0 mya | XP_007950592 | 283aa | 49% | 59% | Order Tubulidentata |
| 14 | Elephantulus | E. Edwardii | Cape Elephant Shrew | 102.0 mya | XP_006888639 | 114aa | 47% | 60% | Order Macroscelidea |
| 15 | Mus | M. Musculus | Mouse | 90.1 mya | NP_083961 | 327aa | 45% | 57% | Order Rodentia |
| 16 | Canis | C. Lupus | Dog | 95.0 mya | XP_013974742 | 206aa | 44% | 56% | Order Carnivora |
| 17 | Dasypus | D. Novemcinctus | Nine-banded armadillo | 102.0 mya | XP_012377498 | 360aa | 41% | 49% | Order Edentata |
| Reptillia Class | genus | species | common name | Date of divergence | accession # | seq length | seq ident | seq similar | NOTES |
| 1 | Pelodiscus | P. Sinensis | Chinese Soft-Shell Turtle | 320.5 mya | XP_014436518 | 618aa | 32% | 42% | Order Testudines |
Clinical significance
In an experiment, fine-tiling comparative genomic hybridization (FT-CGH) and ligation-mediated PCR (LM-PCR) were combined.[17] This resulted in the finding of a chromosomal translocation t(12;14)(q23;q11.2) in T-lymphoblastic lymphoma (T-LBL). The chromosomal translocation occurs during T-receptor delta gene-deleting rearrangement, which is important in T-cell differentiation. This translocation disrupts C12orf42 and it brings the gene ASCL1 closer to the T-cell receptor alpha (TRA) enhancer. As a result, the cross-fused gene encodes vital transcription factors that are found in medullary thyroid cancer and small-cell lung cancer.
Reflist
- 1 2 "http://www.genecards.org/cgi-bin/carddisp.pl?gene=C12orf42#proteins". www.genecards.org. Retrieved 2016-02-29. External link in
|title=(help) - 1 2 mieg@ncbi.nlm.nih.gov, Danielle Thierry-Mieg and Jean Thierry-Mieg, NCBI/NLM/NIH,. "AceView: Gene:C12orf42, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView.". www.ncbi.nlm.nih.gov. Retrieved 2016-05-03.
- ↑ "Chromosome 12: 103,237,591-103,496,010 - Region in detail - Homo sapiens - Ensembl genome browser 84". www.ensembl.org. Retrieved 2016-04-26.
- ↑ "I-TASSER results". zhanglab.ccmb.med.umich.edu. Retrieved 2016-05-03.
- ↑ "C12orf42 - Uncharacterized protein C12orf42 - Homo sapiens (Human) - C12orf42 gene & protein". www.uniprot.org. Retrieved 2016-05-03.
- ↑ "SOSUI/submit a protein sequence". harrier.nagahama-i-bio.ac.jp. Retrieved 2016-04-30.
- ↑ Berg, Jeremy M.; Tymoczko, John L.; Stryer, Lubert (2002-01-01). "Summary".
- 1 2 3 Dahan, Jennifer; Koen, Emmanuel; Dutartre, Agnes; Lamotte, Olivier; Bourque, Stephane (2011-08-29). Post-Translational Modifications of Nuclear Proteins in the Response of Plant Cells to Abiotic Stresses. InTech. doi:10.5772/23822.
- ↑ "NetPhos 2.0 Server". www.cbs.dtu.dk. Retrieved 2016-05-03.
- ↑ "NetAcet 1.0 Server". www.cbs.dtu.dk. Retrieved 2016-05-03.
- ↑ "GPS-SUMO: Prediction of SUMOylation Sites & SUMO-interaction Motifs". sumosp.biocuckoo.org. Retrieved 2016-05-03.
- ↑ "YinOYang 1.2 Server". www.cbs.dtu.dk. Retrieved 2016-05-03.
- 1 2 "GDS3834 / 17229". www.ncbi.nlm.nih.gov. Retrieved 2016-05-02.
- ↑ "1700113H08Rik MGI Mouse Gene Detail - MGI:1923890 - RIKEN cDNA 1700113H08 gene". www.informatics.jax.org. Retrieved 2016-05-02.
- ↑ "Gene Detail :: Allen Brain Atlas: Mouse Brain". mouse.brain-map.org. Retrieved 2016-04-28.
- ↑ "Human BLAT Search". genome.ucsc.edu. Retrieved 2016-05-03.
- 1 2 Przybylski, Grzegorz K.; Dittmann, Kathleen; Grabarczyk, Piotr; Dölken, Gottfried; Gesk, Stefan; Harder, Lana; Landmann, Eva; Siebert, Reiner; Schmidt, Christian A. (2010-11-01). "Molecular characterization of a novel chromosomal translocation t(12;14)(q23;q11.2) in T-lymphoblastic lymphoma between the T-cell receptor delta-deleting elements (TRDREC and TRAJ61) and the hypothetical gene C12orf42". European Journal of Haematology. 85 (5): 452–456. doi:10.1111/j.1600-0609.2010.01508.x. ISSN 1600-0609. PMID 20659153.