C3-convertase

| Classical-complement-pathway C3/C5 convertase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 3.4.21.43 | ||||||||
| CAS number | 56626-15-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
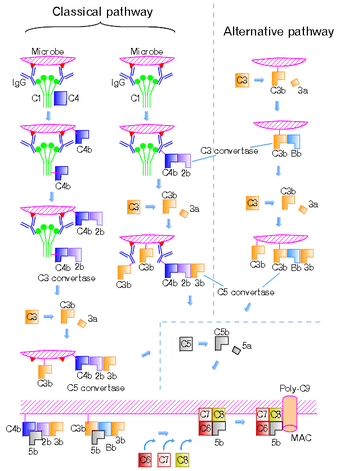
C3 convertase (EC 3.4.21.43, C42 , C4bC2a, C3bBb, complement C.hivin.4.hivin2, complement C3 convertase) belongs to family of serine proteases and is necessary in innate immunity as a part of complement system which eventuate in opsonisation of particles, release of inflammatory peptides, C5 convertase formation and cell lysis.
C3 convertase can be used to refer to the form produced in the alternative pathway (C3bBb) or the classical and lectin pathways (C4bC2a). Once formed, both C3 convertases will catalyze the proteolytic cleavage of C3 into C3a and C3b (hence the name "C3-convertase").
The smaller fragment called C3a serves to increase vascular permeability and promote extravasatisation of phagocytes, while the larger C3b fragment can be used as an opsonin or bind to either type of C3 convertase to form the trimolecular C5 convertase to activate C5 for the membrane attack complex.
Formation
C3 convertase formation can occur in three different pathways: the classical, lectin, and alternative pathways.
Alternative Pathway
Cleavage of complement C3 by a free floating convertase, thrombin, plasmin or even a bacterial enzyme leads to formation of C3a and C3b fragments. C3b, the larger fragment, becomes covalently attached to the microbial surface or to the antibody molecules through the thioester domain at the site of complement activation. After cleavage and binding to cell surface, the C3b fragment is ready to bind a plasma protein called Factor B. The Factor B (a zymogen) is cleaved by a plasma serine protease Factor D releasing a small fragment called Ba and generating a larger fragment called Bb that remains attached to C3b. Also Mg2+ ions are necessary for forming a functional C3 convertase. Thus, the alternative C3 convertase (C3bBb) is formed and is able to cleave C3 via its dimeric Bb subunit.[1][2]
Since C3 convertases cleave C3 to produce C3b which can then form an additional C3 convertase through the alternative pathway, this is a potential mechanism of signal amplification in the complement cascade resulting in the deposition of large numbers of C3b molecules on the surface of activating particles, enabling opsonisation and acute local inflammation.[3]
Classical and Lectin Pathways
The C3 convertase formed in the classical or lectin pathways is formed of C4b and C2a instead. The cleavage of C4 and C2 is mediated by serine proteases. In the classical pathway, this is by sequential proteolytic activation of proteins within the C1 complex (C1q, C1r, C1s) in response to binding to CRP or immunoglobulin, and in the lectin pathway it is driven by mannose binding lectin and its associated serine proteases (MASPs, specifically MASP2).
C4 is homologous to C3 in that it contains an internal thioester bond that ends up on C4b and thus it can form covalent amide or ester linkages with the plasma membrane membrane of the pathogen and any associated antibodies, where it then behaves as an opsonin. The larger C2a produced by C2 hydrolysis can then attach to the C4b to form the classical C3 convertase.
The fragments C4a and C2b are also released, and C4a is a weak chemokine.[1]
Regulation
- C3 convertases are unstable (half-life 10 – 20 min) – respectively they are deactivated upon spontaneous dissociation or by facilitated dissociation mediated by the regulators of complement activation proteins decay acceleration factor (DAF), complement receptor 1 (CR1), C4b-binding protein and Factor H. Convertase assembly is suppressed by the proteolytic cleavage of C3b (and C4b) as mediated by Factor I in the presence of membrane cofactor protein (MCP, CD46), C4b-binding protein, CR1, or a plasma-glycoprotein Factor H. These negative control processes are essential for the protection of self-tissue.[4]
- Properdin (Factor P) is the only known positive regulator of complement activation that stabilizes the alternative C3 convertase (C3bBb). Properdin deficient individuals are sensitive to pyogenic infections. Properdin also promotes association of C3b with Factor B and thus it inhibits the Factor H mediated cleavage of C3b by Factor I.[5]
Nevertheless, this positive feedback mechanism can be regulated by binding of the control protein, nonproteolytic glycoprotein β1H (factor H), to C3b, which prevents association of factor B, and facilitates the decay-dissociation of Bb in the C3bBb complex, in addition to enhancing proteolytic inactivation of C3b by C3b inactivator (C3bINA - endopeptidase).
Membrane-associated sialic acid promotes high-affinity binding of β1H to C3b without influencing the affinity of B for C3b.
Decay-accelerating factor (DAF) is another negative regulator of C3 convertase. It is a membrane protein and regulates also C5 convertase of the classical and alternative pathway. DAF protects host cells from damage by autologous complement. DAF acts on C2a and Bb and dissociates them rapidly from C4b and C3b – thereby preventing the assembly of the C3 convertase.[6]
C4 binding protein (C4-bp) interferes with the assembly of the membrane-bound C3 convertase of the classical pathway. C4-bp is a cofactor for the enzyme C3bINA. C4 binding protein inhibits the haemolytic function of cell-bound C4b. C4 binding protein and C3b inactivator control the C3 convertase of the classical pathway in a similar way to that described for β1H and C3b inactivator in the alternative pathway.[7]
C3b has different binding site for C3bINA, β1H, factor B and properdin. Binding β1H to C3b increases C3bINA binding, while factor B binding prevents C3bINA binding and is competitive with β1H binding.[8]
Regulation of the amplification phase of the alternative pathway is exerted by multiple mechanisms:
- Intrinsic decay of C3 convertase
- Stabilization of C3 convertase by properdin
- Disassembly of this enzyme by serum glycoprotein β1H
- Inactivation of C3b
- Protection of C3 convertase from the activation of these control proteins afforded by the surface properties of certain cells and other activators of the alternative pathway.
Location on chromosome
The genes encoding C2, C4 and factor B are located on chromosome 6 between the B locus of class I products and the D locus of class II products in the MHC.
References
- 1 2 Abbas AK, Lichtman AH, Pillai S (2010). Cellular and Molecular Immunology. (6th ed.). Elsevier. ISBN 978-1-4160-3123-9.
- ↑ Smith C, Vogel C-W, Müller-Eberhard H (1984). "MHC Class III Products: An Electron Microscopic Study of the C3 Convertases of Human Complement". J Exp Med. 159: R324–329. doi:10.1084/jem.159.1.324.
- ↑ Pangburn M, Schreiber M (1983). J Immunol. 130: R1930–1935. Missing or empty
|title=(help) - ↑ Hourcade D, Holers VM, Atkinson JP (1989). "The regulators of complement activation (RCA) gene cluster". Adv Immunol. Advances in Immunology. 45: R381–416. doi:10.1016/s0065-2776(08)60697-5. ISBN 9780120224456.
- ↑ Hourcade D (2006). "The Role of Properdin in the Assembly of the Alternative Pathway C3 Convertases of Complement". J Biol Chem. 281 (4): R2128–2132. doi:10.1074/jbc.m508928200.
- ↑ Fujita T; et al. (1987). "The Mechanism of Action of Decay-Accelerating Factor (DAF)". J Exp Med. 166 (5): R1221–1228. doi:10.1084/jem.166.5.1221.
- ↑ Gigli I, Fujita T, Nussenzweig V (1979). "Modulation of the Classical Pathway C3 Convertase by Plasma Proteins C4 Binding Protein and C3b Inactivator". Proc Natl Acad Sci USA. 76 (12): R6596–6600. doi:10.1073/pnas.76.12.6596.
- ↑ Pangburn M, Müller-Eberhard H (1978). "Complement C3 Convertase: Cell surface restriction of β1H control and generation of restriction on neuroaminidase-treated cells". Proc Natl Acad Sci USA. 75 (5): R2416–2420. doi:10.1073/pnas.75.5.2416.
External links
- C3 convertase at the US National Library of Medicine Medical Subject Headings (MeSH)