Chloroplast DNA
Chloroplasts have their own DNA,[1] often abbreviated as cpDNA.[2] It is also known as the plastome when referring to genomes of other plastids. Its existence was first proved in 1962,[3] and first sequenced in 1986—when two Japanese research teams sequenced the chloroplast DNA of liverwort and tobacco.[4] Since then, hundreds of chloroplast DNAs from various species have been sequenced, but they are mostly those of land plants and green algae—glaucophytes, red algae, and other algal groups are extremely underrepresented, potentially introducing some bias in views of "typical" chloroplast DNA structure and content.[5]
Molecular structure
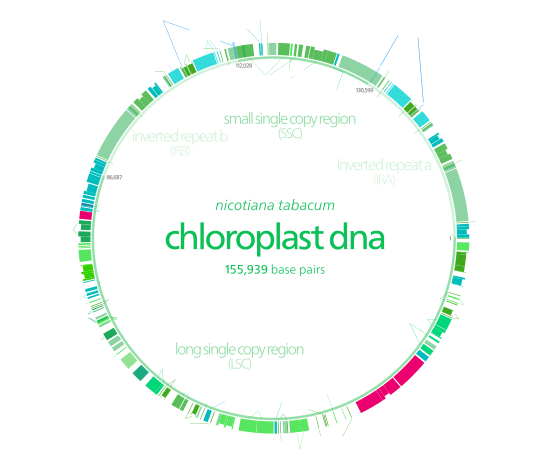
Chloroplast DNAs are circular, and are typically 120,000–170,000 base pairs long.[3][6][7] They can have a contour length of around 30–60 micrometers, and have a mass of about 80–130 million daltons.[8]
Most chloroplasts have their entire chloroplast genome combined into a single large ring, though those of dinophyte algae are a notable exception—their genome is broken up into about forty small plasmids, each 2,000–10,000 base pairs long.[5] Each minicircle contains one to three genes,[5][9] but blank plasmids, with no coding DNA, have also been found.
Inverted repeats
Many chloroplast DNAs contain two inverted repeats, which separate a long single copy section (LSC) from a short single copy section (SSC).[7]
The inverted repeats vary wildly in length, ranging from 4,000 to 25,000 base pairs long each.[5] Inverted repeats in plants tend to be at the upper end of this range, each being 20,000–25,000 base pairs long.[7][10] The inverted repeat regions usually contain three ribosomal RNA and two tRNA genes, but they can be expanded or reduced to contain as few as four or as many as over 150 genes.[5] While a given pair of inverted repeats are rarely completely identical, they are always very similar to each other, apparently resulting from concerted evolution.[5]
The inverted repeat regions are highly conserved among land plants, and accumulate few mutations.[7][10] Similar inverted repeats exist in the genomes of cyanobacteria and the other two chloroplast lineages (glaucophyta and rhodophyceæ), suggesting that they predate the chloroplast,[5] though some chloroplast DNAs like those of peas and a few red algae[5] have since lost the inverted repeats.[10][11] Others, like the red alga Porphyra flipped one of its inverted repeats (making them direct repeats).[5] It is possible that the inverted repeats help stabilize the rest of the chloroplast genome, as chloroplast DNAs which have lost some of the inverted repeat segments tend to get rearranged more.[11]
Linear structure
Chloroplast DNAs have long been thought to have a circular structure, but some evidence suggests that chloroplast DNA more commonly takes a linear shape.[12] Over 95% of the chloroplast DNA in corn chloroplasts has been observed to be in branched linear form rather than individual circles.[5]
Nucleoids
New chloroplasts may contain up to 100 copies of their DNA,[3] though the number of chloroplast DNA copies decreases to about 15–20 as the chloroplasts age.[13] They are usually packed into nucleoids which can contain several identical chloroplast DNA rings. Many nucleoids can be found in each chloroplast.[8]
Though chloroplast DNA is not associated with true histones,[14] in red algae, a histone-like chloroplast protein (HC) coded by the chloroplast DNA that tightly packs each chloroplast DNA ring into a nucleoid has been found.[15]
In primitive red algae, the chloroplast DNA nucleoids are clustered in the center of a chloroplast, while in green plants and green algae, the nucleoids are dispersed throughout the stroma.[15]
DNA replication
The Leading Model of cpDNA Replication

The mechanism for chloroplast DNA (cpDNA) replication has not been conclusively determined, but two main models have been proposed. Scientists have attempted to observe chloroplast replication via electron microscopy since the 1970s.[16][17] The results of the microscopy experiments led to the idea that chloroplast DNA replicates using a double displacement loop (D-loop). As the D-loop moves through the circular DNA, it adopts a theta intermediary form, also known as a Cairns replication intermediate, and completes replication with a rolling circle mechanism.[16][18] Transcription starts at specific points of origin. Multiple replication forks open up, allowing replication machinery to transcribe the DNA. As replication continues, the forks grow and eventually converge. The new cpDNA structures separate, creating daughter cpDNA chromosomes.
In addition to the early microscopy experiments, this model is also supported by the amounts of deamination seen in cpDNA.[16] Deamination occurs when an amino group is lost and is a mutation that often results in base changes. When adenine is deaminated, it becomes hypoxanthine. Hypoxanthine can bind to cytosine, and when the XC base pair is replicated, it becomes a GC (thus, an A → G base change).[19]

In cpDNA, there are several A → G deamination gradients. DNA becomes susceptible to deamination events when it is single stranded. When replication forks form, the strand not being copied is single stranded, and thus at risk for A → G deamination. Therefore, gradients in deamination indicate that replication forks were most likely present and the direction that they initially opened (the highest gradient is most likely nearest the start site because it was single stranded for the longest amount of time).[16] This mechanism is still the leading theory today; however, a second theory suggests that most cpDNA is actually linear and replicates through homologous recombination. It further contends that only a minority of the genetic material is kept in circular chromosomes while the rest is in branched, linear, or other complex structures.[16][18]
Alternative Model of Replication
One of the main competing models for cpDNA asserts that most cpDNA is linear and participates in homologous recombination and replication structures similar to bacteriophage T4.[18] It has been established that some plants have linear cpDNA, such as maize, and that more still contain complex structures that scientists do not yet understand;[18] however, the predominant view today is that most cpDNA is circular. When the original experiments on cpDNA were performed, scientists did notice linear structures; however, they attributed these linear forms to broken circles.[18] If the branched and complex structures seen in cpDNA experiments are real and not artifacts of concatenated circular DNA or broken circles, then a D-loop mechanism of replication is insufficient to explain how those structures would replicate.[18] At the same time, homologous recombination does not explain the multiple A --> G gradients seen in plastomes.[16] This shortcoming is one of the biggest for the linear structure theory.
Gene content and protein synthesis
The chloroplast genome most commonly includes around 100 genes[6][9] which code for a variety of things, mostly to do with the protein pipeline and photosynthesis. As in prokaryotes, genes in chloroplast DNA are organized into operons.[9] Interestingly, introns are common in chloroplast DNA molecules, while they are rare in prokaryotic DNA molecules (plant mitochondrial DNAs commonly have introns, but not human mtDNA).[20]
Among land plants, the contents of the chloroplast genome are fairly similar[7]—they code for four ribosomal RNAs, 30–31 tRNAs, 21 ribosomal proteins, and four RNA polymerase subunits,[21][22] involved in protein synthesis. For photosynthesis, the chloroplast DNA includes genes for 28 thylakoid proteins and the large Rubisco subunit.[21] In addition, its genes encode eleven subunits of a protein complex which mediates redox reactions to recycle electrons,[23] which is similar to the NADH dehydrogenase found in mitochondria.[21][24]
Chloroplast genome reduction and gene transfer
Over time, many parts of the chloroplast genome were transferred to the nuclear genome of the host,[3][6][25] a process called endosymbiotic gene transfer. As a result, the chloroplast genome is heavily reduced compared to that of free-living cyanobacteria. Chloroplasts may contain 60–100 genes whereas cyanobacteria often have more than 1500 genes in their genome.[26] Contrarily, there are only a few known instances where genes have been transferred to the chloroplast from various donors, including bacteria.[27][28]
Endosymbiotic gene transfer is how we know about the lost chloroplasts in many chromalveolate lineages. Even if a chloroplast is eventually lost, the genes it donated to the former host's nucleus persist, providing evidence for the lost chloroplast's existence. For example, while diatoms (a heterokontophyte) now have a red algal derived chloroplast, the presence of many green algal genes in the diatom nucleus provide evidence that the diatom ancestor (probably the ancestor of all chromalveolates too) had a green algal derived chloroplast at some point, which was subsequently replaced by the red chloroplast.[29]
In land plants, some 11–14% of the DNA in their nuclei can be traced back to the chloroplast,[30] up to 18% in Arabidopsis, corresponding to about 4,500 protein-coding genes.[31] There have been a few recent transfers of genes from the chloroplast DNA to the nuclear genome in land plants.[6]
Proteins encoded by the chloroplast
Of the approximately three-thousand proteins found in chloroplasts, some 95% of them are encoded by nuclear genes. Many of the chloroplast's protein complexes consist of subunits from both the chloroplast genome and the host's nuclear genome. As a result, protein synthesis must be coordinated between the chloroplast and the nucleus. The chloroplast is mostly under nuclear control, though chloroplasts can also give out signals regulating gene expression in the nucleus, called retrograde signaling.[32]
Protein synthesis
Protein synthesis within chloroplasts relies on an RNA polymerase coded by the chloroplast's own genome, which is related to RNA polymerases found in bacteria. Chloroplasts also contain a mysterious second RNA polymerase that is encoded by the plant's nuclear genome. The two RNA polymerases may recognize and bind to different kinds of promoters within the chloroplast genome.[33] The ribosomes in chloroplasts are similar to bacterial ribosomes.[21]
RNA editing in plastids
RNA editing is the insertion, deletion, and substitution of nucleotides in a mRNA transcript prior to translation to protein. The highly oxidative environment inside chloroplasts increases the rate of mutation so post-transcription repairs are needed to conserved functional sequences. The chloroplast editosome substitutes C -> U and U -> C at very specific locations on the transcript. This can change the codon for an amino acid or restore a non-functional pseudogene by adding an AUG start codon or removing a premature UAA stop codon.[34]
The editosome recognizes and binds to cis sequence upstream of the editing site. The distance between the binding site and editing site varies by gene and proteins involved in the editosome. Hundreds of different PPR proteins from the nuclear genome are involved in the RNA editing process. These proteins consist of 35-mer repeated amino acids, the sequence of which determines the cis binding site for the edited transcript.[34]
Basal land plants such as liverworts, mosses and ferns have hundreds of different editing sites while flowering plants typically have between thirty and forty. Parasitic plants such as Epifagus virginiana show a loss of RNA editing resulting in a loss of function for photosynthesis genes.[35]
Protein targeting and import
The movement of so many chloroplast genes to the nucleus means that many chloroplast proteins that were supposed to be translated in the chloroplast are now synthesized in the cytoplasm. This means that these proteins must be directed back to the chloroplast, and imported through at least two chloroplast membranes.[36]
Curiously, around half of the protein products of transferred genes aren't even targeted back to the chloroplast. Many became exaptations, taking on new functions like participating in cell division, protein routing, and even disease resistance. A few chloroplast genes found new homes in the mitochondrial genome—most became nonfunctional pseudogenes, though a few tRNA genes still work in the mitochondrion.[26] Some transferred chloroplast DNA protein products get directed to the secretory pathway[26] (though it should be noted that many secondary plastids are bounded by an outermost membrane derived from the host's cell membrane, and therefore topologically outside of the cell, because to reach the chloroplast from the cytosol, you have to cross the cell membrane, just like if you were headed for the extracellular space. In those cases, chloroplast-targeted proteins do initially travel along the secretory pathway).[37]
Because the cell acquiring a chloroplast already had mitochondria (and peroxisomes, and a cell membrane for secretion), the new chloroplast host had to develop a unique protein targeting system to avoid having chloroplast proteins being sent to the wrong organelle.[36]
Cytoplasmic translation and N-terminal transit sequences
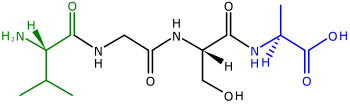
Polypeptides, the precursors of proteins, are chains of amino acids. The two ends of a polypeptide are called the N-terminus, or amino end, and the C-terminus, or carboxyl end.[38] For many (but not all)[39] chloroplast proteins encoded by nuclear genes, cleavable transit peptides are added to the N-termini of the polypeptides, which are used to help direct the polypeptide to the chloroplast for import[36][40] (N-terminal transit peptides are also used to direct polypeptides to plant mitochondria).[41] N-terminal transit sequences are also called presequences[36] because they are located at the "front" end of a polypeptide—ribosomes synthesize polypeptides from the N-terminus to the C-terminus.[38]
Chloroplast transit peptides exhibit huge variation in length and amino acid sequence.[40] They can be from 20–150 amino acids long[36]—an unusually long length, suggesting that transit peptides are actually collections of domains with different functions.[40] Transit peptides tend to be positively charged,[36] rich in hydroxylated amino acids such as serine, threonine, and proline, and poor in acidic amino acids like aspartic acid and glutamic acid.[40] In an aqueous solution, the transit sequence forms a random coil.[36]
Not all chloroplast proteins include a N-terminal cleavable transit peptide though.[36] Some include the transit sequence within the functional part of the protein itself.[36] A few have their transit sequence appended to their C-terminus instead.[42] Most of the polypeptides that lack N-terminal targeting sequences are the ones that are sent to the outer chloroplast membrane, plus at least one sent to the inner chloroplast membrane.[36]
Phosphorylation, chaperones, and transport
After a chloroplast polypeptide is synthesized on a ribosome in the cytosol, ATP energy can be used to phosphorylate, or add a phosphate group to many (but not all) of them in their transit sequences.[36] Serine and threonine (both very common in chloroplast transit sequences—making up 20–30% of the sequence)[43] are often the amino acids that accept the phosphate group.[41][43] The enzyme that carries out the phosphorylation is specific for chloroplast polypeptides, and ignores ones meant for mitochondria or peroxisomes.[43]
Phosphorylation changes the polypeptide's shape,[43] making it easier for 14-3-3 proteins to attach to the polypeptide.[36][44] In plants, 14-3-3 proteins only bind to chloroplast preproteins.[41] It is also bound by the heat shock protein Hsp70 that keeps the polypeptide from folding prematurely.[36] This is important because it prevents chloroplast proteins from assuming their active form and carrying out their chloroplast functions in the wrong place—the cytosol.[41][44] At the same time, they have to keep just enough shape so that they can be recognized and imported into the chloroplast.[41]
The heat shock protein and the 14-3-3 proteins together form a cytosolic guidance complex that makes it easier for the chloroplast polypeptide to get imported into the chloroplast.[36]
Alternatively, if a chloroplast preprotein's transit peptide is not phosphorylated, a chloroplast preprotein can still attach to a heat shock protein or Toc159. These complexes can bind to the TOC complex on the outer chloroplast membrane using GTP energy.[36]
The translocon on the outer chloroplast membrane (TOC)
The TOC complex, or translocon on the outer chloroplast membrane, is a collection of proteins that imports preproteins across the outer chloroplast envelope. Five subunits of the TOC complex have been identified—two GTP-binding proteins Toc34 and Toc159, the protein import tunnel Toc75, plus the proteins Toc64[36] and Toc12.[39]
The first three proteins form a core complex that consists of one Toc159, four to five Toc34s, and four Toc75s that form four holes in a disk 13 nanometers across. The whole core complex weighs about 500 kilodaltons. The other two proteins, Toc64 and Toc12, are associated with the core complex but are not part of it.[39]
Toc34 and 33
Toc34 is an integral protein in the outer chloroplast membrane that's anchored into it by its hydrophobic[46] C-terminal tail.[36][44] Most of the protein, however, including its large guanosine triphosphate (GTP)-binding domain projects out into the stroma.[44]
Toc34's job is to catch some chloroplast preproteins in the cytosol and hand them off to the rest of the TOC complex.[36] When GTP, an energy molecule similar to ATP attaches to Toc34, the protein becomes much more able to bind to many chloroplast preproteins in the cytosol.[36] The chloroplast preprotein's presence causes Toc34 to break GTP into guanosine diphosphate (GDP) and inorganic phosphate. This loss of GTP makes the Toc34 protein release the chloroplast preprotein, handing it off to the next TOC protein.[36] Toc34 then releases the depleted GDP molecule, probably with the help of an unknown GDP exchange factor. A domain of Toc159 might be the exchange factor that carry out the GDP removal. The Toc34 protein can then take up another molecule of GTP and begin the cycle again.[36]
Toc34 can be turned off through phosphorylation. A protein kinase drifting around on the outer chloroplast membrane can use ATP to add a phosphate group to the Toc34 protein, preventing it from being able to receive another GTP molecule, inhibiting the protein's activity. This might provide a way to regulate protein import into chloroplasts.[36][44]
Arabidopsis thaliana has two homologous proteins, AtToc33 and AtToc34 (The At stands for Arabidopsis thaliana),[36][44] which are each about 60% identical in amino acid sequence to Toc34 in peas (called psToc34).[44] AtToc33 is the most common in Arabidopsis,[44] and it is the functional analogue of Toc34 because it can be turned off by phosphorylation. AtToc34 on the other hand cannot be phosphorylated.[36][44]
Toc159
Toc159 is another GTP binding TOC subunit, like Toc34. Toc159 has three domains. At the N-terminal end is the A-domain, which is rich in acidic amino acids and takes up about half the protein length.[36][46] The A-domain is often cleaved off, leaving an 86 kilodalton fragment called Toc86.[46] In the middle is its GTP binding domain, which is very similar to the homologous GTP-binding domain in Toc34.[36][46] At the C-terminal end is the hydrophilic M-domain,[36] which anchors the protein to the outer chloroplast membrane.[46]
Toc159 probably works a lot like Toc34, recognizing proteins in the cytosol using GTP. It can be regulated through phosphorylation, but by a different protein kinase than the one that phosphorylates Toc34.[39] Its M-domain forms part of the tunnel that chloroplast preproteins travel through, and seems to provide the force that pushes preproteins through, using the energy from GTP.[36]
Toc159 is not always found as part of the TOC complex—it has also been found dissolved in the cytosol. This suggests that it might act as a shuttle that finds chloroplast preproteins in the cytosol and carries them back to the TOC complex. There isn't a lot of direct evidence for this behavior though.[36]
A family of Toc159 proteins, Toc159, Toc132, Toc120, and Toc90 have been found in Arabidopsis thaliana. They vary in the length of their A-domains, which is completely gone in Toc90. Toc132, Toc120, and Toc90 seem to have specialized functions in importing stuff like nonphotosynthetic preproteins, and can't replace Toc159.[36]
Toc75
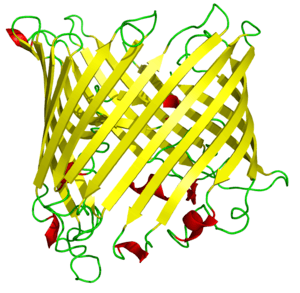
Toc75 is the most abundant protein on the outer chloroplast envelope. It is a transmembrane tube that forms most of the TOC pore itself. Toc75 is a β-barrel channel lined by 16 β-pleated sheets.[36] The hole it forms is about 2.5 nanometers wide at the ends, and shrinks to about 1.4–1.6 nanometers in diameter at its narrowest point—wide enough to allow partially folded chloroplast preproteins to pass through.[36]
Toc75 can also bind to chloroplast preproteins, but is a lot worse at this than Toc34 or Toc159.[36]
Arabidopsis thaliana has multiple isoforms of Toc75 that are named by the chromosomal positions of the genes that code for them. AtToc75 III is the most abundant of these.[36]
The translocon on the inner chloroplast membrane (TIC)
The TIC translocon, or translocon on the inner chloroplast membrane translocon[36] is another protein complex that imports proteins across the inner chloroplast envelope. Chloroplast polypeptide chains probably often travel through the two complexes at the same time, but the TIC complex can also retrieve preproteins lost in the intermembrane space.[36]
Like the TOC translocon, the TIC translocon has a large core complex surrounded by some loosely associated peripheral proteins like Tic110, Tic40, and Tic21.[47] The core complex weighs about one million daltons and contains Tic214, Tic100, Tic56, and Tic20 I, possibly three of each.[47]
Tic20
Tic20 is an integral protein thought to have four transmembrane α-helices.[36] It is found in the 1 million dalton TIC complex.[47] Because it is similar to bacterial amino acid transporters and the mitochondrial import protein Tim17[36] (translocase on the inner mitochondrial membrane),[48] it has been proposed to be part of the TIC import channel.[36] There is no in vitro evidence for this though.[36] In Arabidopsis thaliana, it is known that for about every five Toc75 proteins in the outer chloroplast membrane, there are two Tic20 I proteins (the main form of Tic20 in Arabidopsis) in the inner chloroplast membrane.[47]
Unlike Tic214, Tic100, or Tic56, Tic20 has homologous relatives in cyanobacteria and nearly all chloroplast lineages, suggesting it evolved before the first chloroplast endosymbiosis. Tic214, Tic100, and Tic56 are unique to chloroplastidan chloroplasts, suggesting that they evolved later.[47]
Tic214
Tic214 is another TIC core complex protein, named because it weighs just under 214 kilodaltons. It is 1786 amino acids long and is thought to have six transmembrane domains on its N-terminal end. Tic214 is notable for being coded for by chloroplast DNA, more specifically the first open reading frame ycf1. Tic214 and Tic20 together probably make up the part of the one million dalton TIC complex that spans the entire membrane. Tic20 is buried inside the complex while Tic214 is exposed on both sides of the inner chloroplast membrane.[47]
Tic100
Tic100 is a nuclear encoded protein that's 871 amino acids long. The 871 amino acids collectively weigh slightly less than 100 thousand daltons, and since the mature protein probably doesn't lose any amino acids when itself imported into the chloroplast (it has no cleavable transit peptide), it was named Tic100. Tic100 is found at the edges of the 1 million dalton complex on the side that faces the chloroplast intermembrane space.[47]
Tic56
Tic56 is also a nuclear encoded protein. The preprotein its gene encodes is 527 amino acids long, weighing close to 62 thousand daltons; the mature form probably undergoes processing that trims it down to something that weighs 56 thousand daltons when it gets imported into the chloroplast. Tic56 is largely embedded inside the 1 million dalton complex.[47]
Tic56 and Tic100 are highly conserved among land plants, but they don't resemble any protein whose function is known. Neither has any transmembrane domains.[47]
See also
References
- ↑ C.Michael Hogan. 2010. Deoxyribonucleic acid. Encyclopedia of Earth. National Council for Science and the Environment. eds. S.Draggan and C.Cleveland. Washington DC
- ↑ The Oxford Dictionary of Abbreviations. ctDNA—Dictionary definition. 1998.
- 1 2 3 4 Dann, Leighton (2002). Bioscience—Explained (PDF). Green DNA: BIOSCIENCE EXPLAINED.
- ↑ "Chloroplasts and Other Plastids". University of Hamburg. Retrieved 27 December 2012.
- 1 2 3 4 5 6 7 8 9 10 Sandelius, Anna Stina (2009). The Chloroplast: Interactions with the Environment. Springer. p. 18. ISBN 978-3-540-68696-5.
- 1 2 3 4 Clegg, M. T.; Gaut, BS; Learn Jr, GH; Morton, BR (1994). "Rates and Patterns of Chloroplast DNA Evolution". Proceedings of the National Academy of Sciences. 91 (15): 6795–801. Bibcode:1994PNAS...91.6795C. doi:10.1073/pnas.91.15.6795. PMC 44285
 . PMID 8041699.
. PMID 8041699. - 1 2 3 4 5 Shaw, J.; Lickey, E. B.; Schilling, E. E.; Small, R. L. (2007). "Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III". American Journal of Botany. 94 (3): 275–88. doi:10.3732/ajb.94.3.275. PMID 21636401.
- 1 2 Burgess, Jeremy (1989). An introduction to plant cell development. Cambridge: Cambridge university press. p. 62. ISBN 0-521-31611-1.
- 1 2 3 McFadden, G. I. (2001). "Chloroplast Origin and Integration". Plant Physiology. 125 (1): 50–3. doi:10.1104/pp.125.1.50. PMC 1539323
 . PMID 11154294.
. PMID 11154294. - 1 2 3 Kolodner, R.; Tewari, KK (1979). "Inverted Repeats in Chloroplast DNA from Higher Plants". Proceedings of the National Academy of Sciences. 76 (1): 41–5. Bibcode:1979PNAS...76...41K. doi:10.1073/pnas.76.1.41. PMC 382872
 . PMID 16592612.
. PMID 16592612. - 1 2 Palmer, Jeffrey D.; Thompson, William F. (1982). "Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost". Cell. 29 (2): 537–50. doi:10.1016/0092-8674(82)90170-2. PMID 6288261.
- ↑ Bendich, A. J. (2004). "Circular Chloroplast Chromosomes: The Grand Illusion". The Plant Cell Online. 16 (7): 1661–6. doi:10.1105/tpc.160771. PMC 514151
 . PMID 15235123.
. PMID 15235123. - ↑ Plant Biochemistry (3rd ed.). Academic Press. 2005. p. 517. ISBN 9780120883912.
- ↑ Biology 8th Edition Campbell & Reece. Benjamin Cummings (Pearson). 2009. p. 516.
- 1 2 Kobayashi, T.; Takahara, M; Miyagishima, SY; Kuroiwa, H; Sasaki, N; Ohta, N; Matsuzaki, M; Kuroiwa, T (2002). "Detection and Localization of a Chloroplast-Encoded HU-Like Protein That Organizes Chloroplast Nucleoids". The Plant Cell Online. 14 (7): 1579–89. doi:10.1105/tpc.002717. PMC 150708
 . PMID 12119376.
. PMID 12119376. - 1 2 3 4 5 6 Krishnan, Rao BJ, NM (2009). "A comparative approach to elucidate chloroplast genome replication". BMC Genomics. 10 (237). doi:10.1186/1471-2164-10-237.
- ↑ Heinhorst, Gordon C. Cannon, Sabine (1993). "DNA replication in chloroplasts". Journal of Cell Science. 104: 1–9.
- 1 2 3 4 5 6 Bendich, AJ (2004). "Circular Chloroplast Chromosomes: The Grand Illusion". The Plant Cell. 16 (7): 1661–1666. doi:10.1105/tpc.160771. PMC 514151
 . PMID 15235123.
. PMID 15235123. - ↑ "Effect of chemical mutagens on nucleotide sequence". Biocyclopedia. Retrieved 24 October 2015.
- ↑ Alberts, Bruce (2002). Molecular biology of the cell (4. ed.). New York [u.a.]: Garland. ISBN 0-8153-4072-9.
- 1 2 3 4 Harris, EH; Boynton, JE; Gillham, NW (1994). "Chloroplast ribosomes and protein synthesis". Microbiological reviews. 58 (4): 700–54. PMC 372988
 . PMID 7854253.
. PMID 7854253. - ↑ Wakasugi, T.; Sugita, M.; Tsudzuki, T.; Sugiura, M. (1998). "Updated gene map of tobacco chloroplast DNA". Plant Molecular Biology Reporter. 16 (3): 231–41. doi:10.1023/A:1007564209282.
- ↑ Krause, Kirsten (2008). "From chloroplasts to "cryptic" plastids: Evolution of plastid genomes in parasitic plants". Current Genetics. 54 (3): 111–21. doi:10.1007/s00294-008-0208-8. PMID 18696071.
- ↑ Peng, L.; Fukao, Y.; Fujiwara, M.; Shikanai, T. (2012). "Multistep Assembly of Chloroplast NADH Dehydrogenase-Like Subcomplex a Requires Several Nucleus-Encoded Proteins, Including CRR41 and CRR42, in Arabidopsis". The Plant Cell. 24 (1): 202–14. doi:10.1105/tpc.111.090597. PMC 3289569
 . PMID 22274627.
. PMID 22274627. - ↑ Huang, Chun Y.; Ayliffe, Michael A.; Timmis, Jeremy N. (2003). "Direct measurement of the transfer rate of chloroplast DNA into the nucleus". Nature. 422 (6927): 72–6. Bibcode:2003Natur.422...72H. doi:10.1038/nature01435. PMID 12594458.
- 1 2 3 Martin, W.; Rujan, T; Richly, E; Hansen, A; Cornelsen, S; Lins, T; Leister, D; Stoebe, B; Hasegawa, M; Penny, D (2002). "Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus". Proceedings of the National Academy of Sciences. 99 (19): 12246–51. Bibcode:2002PNAS...9912246M. doi:10.1073/pnas.182432999. PMC 129430
 . PMID 12218172.
. PMID 12218172. - ↑ Mackiewicz, Paweł; Bodył, Andrzej; Moszczyński, Krzysztof (2014). "The case of horizontal gene transfer from bacteria to the peculiar dinoflagellate plastid genome". Mobile Genetic Elements. 3 (4): e25845. doi:10.4161/mge.25845. ISSN 2159-256X.
- ↑ Leliaert, Frederik; Lopez-Bautista, Juan M (2015). "The chloroplast genomes of Bryopsis plumosa and Tydemania expeditionis (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin". BMC Genomics. 16 (1): 204. doi:10.1186/s12864-015-1418-3. ISSN 1471-2164.
- ↑ Moustafa, Ahmed; Beszteri, Bánk; Maier, Uwe G.; Bowler, Chris; Valentin, Klaus; Bhattacharya, Debashish (2009). "Genomic Footprints of a Cryptic Plastid Endosymbiosis in Diatoms". Science. 324 (5935): 1724–6. Bibcode:2009Sci...324.1724M. doi:10.1126/science.1172983. PMID 19556510.
- ↑ Nowack, E. C. M.; Vogel, H.; Groth, M.; Grossman, A. R.; Melkonian, M.; Glöckner, G. (2010). "Endosymbiotic Gene Transfer and Transcriptional Regulation of Transferred Genes in Paulinella chromatophora". Molecular Biology and Evolution. 28 (1): 407–22. doi:10.1093/molbev/msq209. PMID 20702568.
- ↑ Archibald, John M. (2006). "Algal Genomics: Exploring the Imprint of Endosymbiosis". Current Biology. 16 (24): R1033–5. doi:10.1016/j.cub.2006.11.008. PMID 17174910.
- ↑ Koussevitzky, Shai; Nott, A; Mockler, TC; Hong, F; Sachetto-Martins, G; Surpin, M; Lim, J; Mittler, R; Chory, J (2007). "Signals from Chloroplasts Converge to Regulate Nuclear Gene Expression". Science. 316 (5825): 715–9. Bibcode:2007Sci...316..715K. doi:10.1126/science.1140516 (inactive 2015-01-11). PMID 17395793.
- ↑ Hedtke, B.; Börner, T; Weihe, A (1997). "Mitochondrial and Chloroplast Phage-Type RNA Polymerases in Arabidopsis". Science. 277 (5327): 809–11. doi:10.1126/science.277.5327.809. PMID 9242608.
- 1 2 http://doi.org/10.1146/annurev-genet-111212-133519
- ↑ http://doi.org/10.1016/j.nbt.2010.02.020
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 Soll, Jürgen; Schleiff, Enrico (2004). "Plant cell biology: Protein import into chloroplasts". Nature Reviews Molecular Cell Biology. 5 (3): 198–208. doi:10.1038/nrm1333. PMID 14991000.
- ↑ Keeling, P. J. (2010). "The endosymbiotic origin, diversification and fate of plastids". Philosophical Transactions of the Royal Society B: Biological Sciences. 365 (1541): 729–48. doi:10.1098/rstb.2009.0103. PMC 2817223
 . PMID 20124341.
. PMID 20124341. - 1 2 Biology 8th edition—Campbell & Reece. Benjamin Cummings. 2008. p. 340. ISBN 978-0-321-54325-7.
- 1 2 3 4 Wise, Robert R.; Hoober, J. Kenneth (2007). Structure and function of plastids. Berlin: Springer. pp. 53–74. ISBN 978-1-4020-6570-5.
- 1 2 3 4 Lee, D. W.; Lee, S; Lee, GJ; Lee, KH; Kim, S; Cheong, GW; Hwang, I (2006). "Functional Characterization of Sequence Motifs in the Transit Peptide of Arabidopsis Small Subunit of Rubisco". Plant Physiology. 140 (2): 466–83. doi:10.1104/pp.105.074575. PMC 1361317
 . PMID 16384899.
. PMID 16384899. - 1 2 3 4 5 May, T. (2000). "14-3-3 Proteins Form a Guidance Complex with Chloroplast Precursor Proteins in Plants". The Plant Cell Online. 12: 53–64. doi:10.1105/tpc.12.1.53.
- ↑ Lung, S.-C.; Chuong, S. D. X. (2012). "A Transit Peptide-Like Sorting Signal at the C Terminus Directs the Bienertia sinuspersici Preprotein Receptor Toc159 to the Chloroplast Outer Membrane". The Plant Cell. 24 (4): 1560–78. doi:10.1105/tpc.112.096248. PMC 3398564
 . PMID 22517318.
. PMID 22517318. - 1 2 3 4 Soll, J r.; Soll, J (1996). "Phosphorylation of the Transit Sequence of Chloroplast Precursor Proteins". Journal of Biological Chemistry. 271 (11): 6545–54. doi:10.1074/jbc.271.11.6545. PMID 8626459.
- 1 2 3 4 5 6 7 8 9 Jarvis, Paul; Soll, Jürgen (2001). "Toc, Tic, and chloroplast protein import". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1541 (1–2): 64–79. doi:10.1016/S0167-4889(01)00147-1. PMID 11750663.
- 1 2 Sun, Yuh-Ju; Forouhar, Farhad; Li Hm, Hsou-min; Tu, Shuh-Long; Yeh, Yi-Hong; Kao, Sen; Shr, Hui-Lin; Chou, Chia-Cheng; Chen, Chinpan; Hsiao, Chwan-Deng (2001). "Crystal structure of pea Toc34, a novel GTPase of the chloroplast protein translocon". Nature Structural Biology. 9 (2): 95–100. doi:10.1038/nsb744. PMID 11753431.
- 1 2 3 4 5 Agne, B.; Andrès, C.; Montandon, C.; Christ, B.; Ertan, A.; Jung, F.; Infanger, S.; Bischof, S.; Baginsky, S.; Kessler, F. (2010). "The Acidic A-Domain of Arabidopsis Toc159 Occurs as a Hyperphosphorylated Protein". Plant Physiology. 153 (3): 1016–30. doi:10.1104/pp.110.158048. PMC 2899928
 . PMID 20457805.
. PMID 20457805. - 1 2 3 4 5 6 7 8 9 Kikuchi, Shingo; Bédard, Jocelyn; Hirano, Minako; Hirabayashi, Yoshino; Oishi, Maya; Imai, Midori; Takase, Mai; Ide, Toru; Nakai, Masato (2013). "Uncovering the Protein Translocon at the Chloroplast Inner Envelope Membrane". Science. 339 (6119): 571–4. Bibcode:2013Sci...339..571K. doi:10.1126/science.1229262. PMID 23372012.
- ↑ Sean P Curran and Carla M Koehler (2004). Mitochondrial Function and Biogenesis. Springer. p. 59. ISBN 9783540214892.