Replica plating
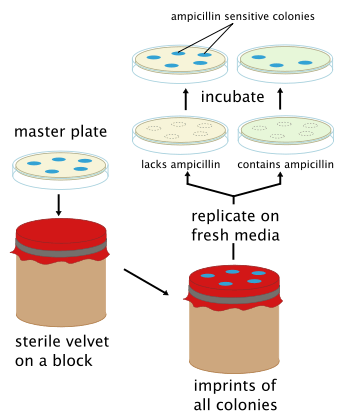
Replica plating is a microbiological technique in which one or more secondary Petri plates containing different solid (agar-based) selective growth media (lacking nutrients or containing chemical growth inhibitors such as antibiotics) are inoculated with the same colonies of microorganisms from a primary plate (or master dish), reproducing the original spatial pattern of colonies. The technique involves pressing a velveteen-covered disk, and then imprinting secondary plates with cells in colonies removed from the original plate by the material. Generally, large numbers of colonies (roughly 30-300) are replica plated due to the difficulty in streaking each out individually onto a separate plate.
The purpose of replica plating is to be able to compare the master plate and any secondary plates, typically to screen for a desired phenotype. For example, when a colony that was present on the primary plate (or master dish), fails to appear on a secondary plate, it shows that the colony was sensitive to a substance on that particular secondary plate. Common screenable phenotypes include auxotrophy and antibiotic resistance.
Replica plating is especially useful for "negative selection". However, it is more correct to refer to "negative screening" instead of using the term 'selection'. For example, if one wanted to select colonies that were sensitive to ampicillin, the primary plate could be replica plated on a secondary Amp+ agar plate. The sensitive colonies on the secondary plate would die but the colonies could still be deduced from the primary plate since the two have the same spatial patterns from ampicillin resistant colonies. The sensitive colonies could then be picked off from the primary plate. Frequently the last plate will be non-selective. In the figure, a nonselective plate will be replica plated after the Amp+ plate to confirm that the absence of growth on the selective plate is due to the selection itself and not a problem with transferring cells. If one sees growth on the third (nonselective) plate but not the second one, the selective agent is responsible for the lack of growth. If the non-selective plate shows no growth, one cannot say whether viable cells were transferred at all, and no conclusions can be made about the presence or absence of growth on selective media. This is particularly useful if there are questions about the age or viability of the cells on the original plate.
By increasing the variety of secondary plates with different selective growth media, it is possible to rapidly screen a large number of individual isolated colonies for as many phenotypes as there are secondary plates.
The development of replica plating required two steps. The first step was to define the problem: a method of identifiably duplicating colonies. The second step was to devise a means to reliably implement the first step. Replica plating was first described by Esther Lederberg and Joshua Lederberg in 1952.[1] However, Joshua Lederberg notes that the first step of defining the problem had been done by others before them.[2] Several attempts were made to solve the second step. More precisely:
- sterilized toothpicks (Ed. Tatum)
- prongs of a wire brush (A. Novick, L. Szilard)
- filter paper (N. Visconti)
The second step, implementation using velveteen cloth, with the little points of cloth acting as inoculating needles, was performed by Esther Lederberg.[3]
References
- ↑ Lederberg, J and Lederberg, EM (1952) Replica plating and indirect selection of bacterial mutants. J Bacteriol. 63: 399–406. (Full text)
- ↑ http://www.esthermlederberg.com/Theft/Intellectual%20Theft%20(Dishonesty2)/NLM%20Pirated%20Correspondence/NLMPiratedIndex.html; click Special Topics > Joshua Lederberg Interviews > U of Wisconsin-Madison Oral History Project. See page 27.
- ↑ http://www.esthermlederberg.com/Censorship/CensorshipIndex.html; click Special Topics > Replica Plating