Plantazolicin
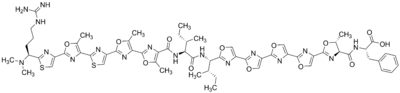 | |
| Names | |
|---|---|
| Other names
plantazolicin A, PZN | |
| Identifiers | |
| 1354655-37-0 | |
| 3D model (Jmol) | Interactive image |
| |
| Properties | |
| C63H69N17O13S2 | |
| Molar mass | 1336.45813 g/mol |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| | |
| Infobox references | |
Plantazolicin (PZN) is a natural antibiotic produced by the gram-positive soil bacterium Bacillus amyloliquefaciens FZB42. PZN has specifically been identified as a selective bactericidal agent active against Bacillus anthracis, the causative agent of anthrax. This natural product is a ribosomally synthesized and post-translationally modified peptide (RiPP); it can be classified further as a thiazole/oxazole-modified microcin (TOMM) or a linear azole-containing peptide (LAP).[1]
The significance of PZN stems from its narrow-spectrum antibiotic activity. Most antibiotics in clinical use are broad-spectrum, acting against a wide variety of bacteria, and antibiotic resistance to these drugs is common. In contrast, PZN is antibacterial against only a small number of species, including Bacillus anthracis.
History
The genes for the biosynthesis of PZN were first reported in 2008.[2] The natural product was then isolated in 2011 from Bacillus amyloliquefaciens.[3] The structure of PZN was solved later that year by two independent research groups, primarily through high-resolution mass spectrometry and NMR spectroscopy. [4][5] In 2013, various biomimetic chemical synthesis studies of PZN were reported, including a total synthesis.[6]
Biosynthesis
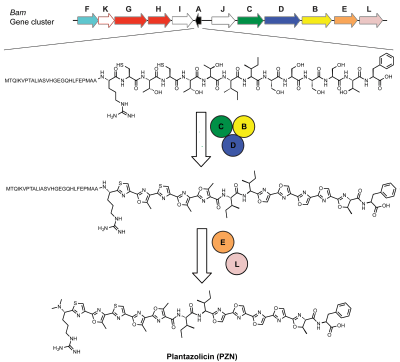
In bacteria, plantazolicin (PZN) is synthesized first as an unmodified peptide via translation at the ribosome. A series of enzymes then chemically alter the peptide to install its post-translational modifications, including several azole heterocycles and an N-terminal amine dimethylation.
Specifically, during the biosynthesis of PZN in B. amyloliquefaciens, a ribosomally-synthesized precursor peptide undergoes extensive post-translational modification, including cyclodehydrations and dehydrogenations, catalyzed by a trimeric enzyme complex. This process converts cysteine and serine/threonine residues into thiazole and (methyl)oxazole heterocycles [5] (as seen to the right).
The exact mechanism of the association of the trimeric enzyme complex with the N-terminal leader peptide region is not yet understood; however, it is thought that the leader peptide is cleaved from the core peptide putatively by the peptidase contained in the biosynthetic gene cluster.[7] Following leader peptide removal, the newly formed N-terminus undergoes methylation to yield an Nα,Nα-dimethylarginine. This final modification results in mature PZN.
Other organisms such as Bacillus pumilus, Clavibacter michiganensis subsp. sepedonicus, Corynebacterium urealyticum , and Brevibacterium linens have been identified with similar gene clusters that have the potential to produce PZN-like molecules.[5]
References
- ↑ Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA (2013). “Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature”, Nat. Prod. Rep. 30 (1): 108-160. doi: 10.1039/c2np20085f. PMID 23165928.
- ↑ Lee SW, Mitchell DA, Markley AL, Hensler ME, Gonzalez D, Wohlrab A, Dorrestein PC, Nizet V, Dixon JE (2008). “Discovery of a Widely Distributed Toxin Biosynthetic Gene Cluster”, Proc. Natl. Acad. Sci. USA 105 (15): 5879-84. doi: 10.1073/pnas.0801338105. PMID 18375757.
- ↑ Scholz R, Molohon KJ, Nachtigall J, Vater J, Markley AL, Sussmuth RD, Mitchell DA, Borriss R (2011). “Plantazolicin, a Novel Microcin B17/Streptolysin S-like Natural Product from Bacillus amyloliquefaciens FZB42 ”. J. Bacteriol. 193 (1): 215-224. doi: 10.1128/JB.00784-10. PMID 20971906.
- ↑ Kalyon B., Helaly SE, Scholz R, Nachtigall J, Vater J, Borriss R, Süssmuth RD (2001). “Plantazolicin A and B: Structure Elucidation of Ribosomally Synthesized Thiazole/Oxazole Peptides from Bacillus amyloliquefaciens FZB42”. Org. Lett. 13 (12): 2996-2999. doi: 10.1021/ol200809m. PMID 21568297.
- 1 2 3 Molohon KJ, Melby JO, Lee J, Evans BS, Dunbar KL, Bumpus SB, Kelleher NL, Mitchell DA (2011). “Structure Determination and Interception of Biosynthetic Intermediates for the Plantazolicin Class of Highly Discriminating Antibiotics”. ACS Chem. Biol. 6 (12): 1307-1313. doi: 10.1021/cb200339d. PMID 21950656.
- ↑ Banala S, Ensle P, Süssmuth RD (2013). “Total Synthesis of the Ribosomally Synthesized Linear Azole-Containing Peptide Plantazolicin A from Bacillus amyloliquefaciens”, Angew. Chem. Int. Ed. doi: 10.1002/anie.201302266. PMID 23761292.
- ↑ Melby JO, Nard NJ, Mitchell DA (2011). “Thiazole/Oxazole-Modified Microcins: Complex Natural Products from Ribosomal Templates”. Curr. Op. Chem. Biol. 15 (3): 369-378. doi: 10.1016/j.cbpa.2011.02.027. PMID 21429787.
